 |
|
Systems Biology and Network Analysis |
|
 |
FunCoup
Explore comprehensive gene/protein networks of functional coupling.
|
|
 |
FunCoup Cytoscape app
Explore FunCoup's comprehensive networks with the power of Cytoscape.
|
|
 |
PathBIX
Pathway analysis using network crosstalk using ANUBIX.
|
|
 |
PathwAX
Pathway analysis using network crosstalk using BinoX.
|
|
 |
Online GRN Benchmark
Benchmark your own inferred gene regulatory networks and see results of public methods.
|
|
 |
CancerGRN
Explore cancer gene regulatory networks.
|
|
 |
MaxLink
Network-based Identification of Genes Related to a Seed Gene Set.
|
|
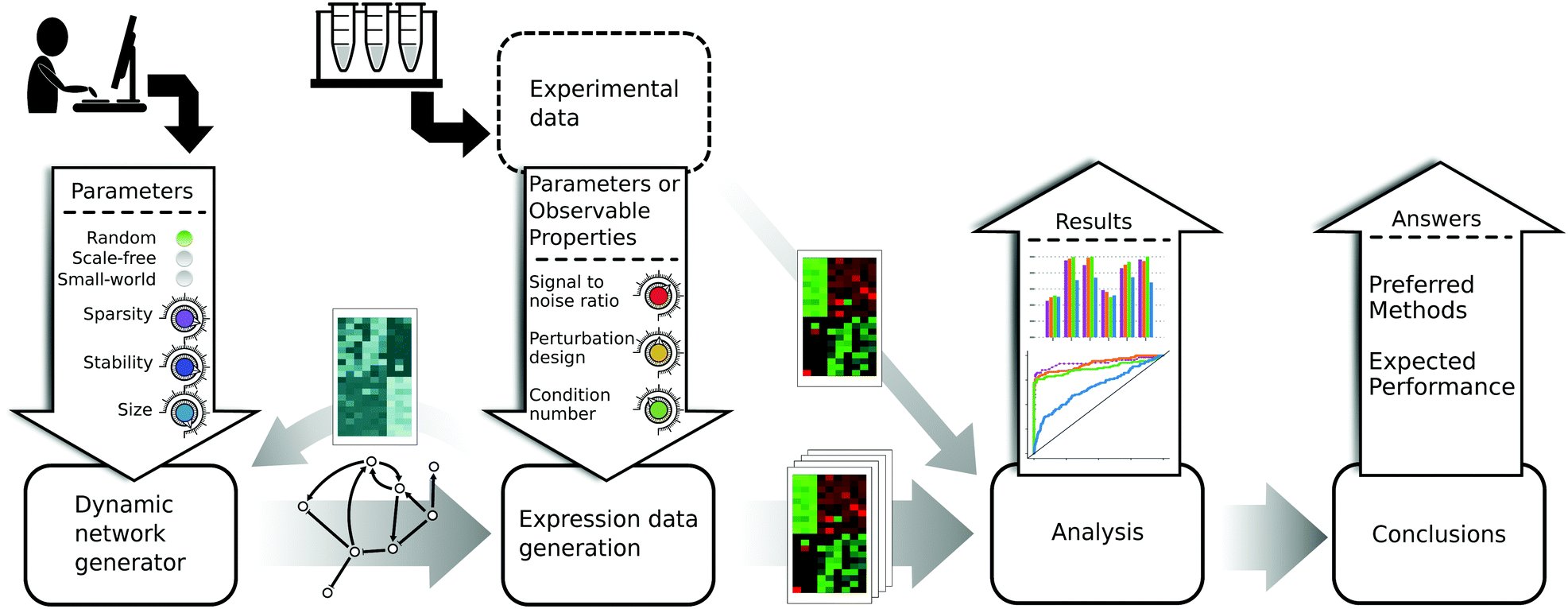 |
GeneSpider
GeneSPIDER Matlab toolbox for generation and analysis of gene regulatory networks and data.
|
|
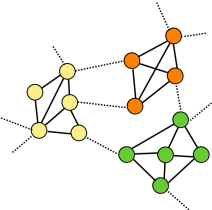 |
MGclus
Network clustering employing shared neighbors.
|
|
 |
UniDomInt
A comprehensive database of domain-domain interactions.
|
 |
|
Ortholog Detection and Analysis |
|
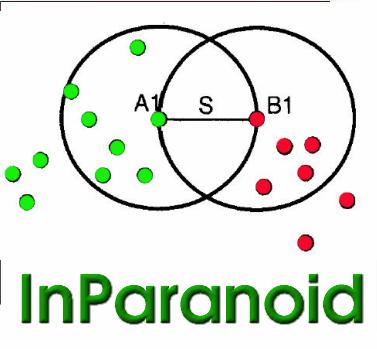 |
InParanoiDB
A database of orthologs between 640 species, including inparalogs for both protein domains and full-length proteins.
|
|
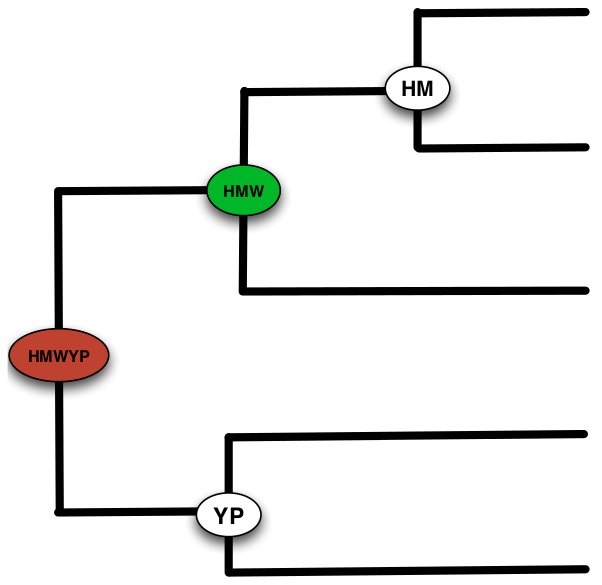 |
Hieranoid
Hieranoid is an orthology inference method using a hierarchical
approach. Hieranoid performs pairwise orthology analysis using
InParanoid at each node in a guide tree as it progresses from its
leaves to the root. This concept reduces the total runtime complexity
from a quadratic to a linear function of the number of species.
|
|
 |
HieranoiDB
An online database of Hieranoid orthologs.
|
|
 |
OrthoXML and SeqXML
XML standards for orthology and sequence information.
|
|
 |
Orthodisease
A database of disease gene orthologs.
|
|
 |
MultiParanoid
A multi-species version of InParanoid. This is no longer supported - use Hieranoid instead.
|
|
 |
Humanoid
Human ortholog groups and functional shift analysis of subfamilies in these.
|
|
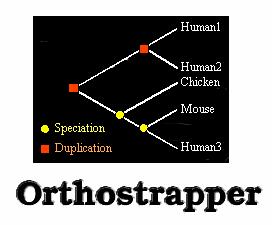 |
Orthostrapper
Extracts orthology support values from trees. It browses a sequence tree for orthologous relationships between two species.
|
|
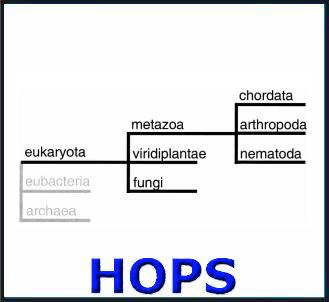 |
HOPS
Hierarchical grouping of Orthologous and Paralagous sequences. This is a database of Orthologous protein domains in Pfam, where the Orthology is inferred in a hierarchic system of phylogenetic subgroups using ortholog bootstrapping.
|
 |
|
Protein Function / Transmembrane Topology / Structure Analysis |
|
 |
MetaTM
A consensus method for predicting transmembrane topology.
|
|
 |
Phobius
A combined transmembrane topology and signal peptide predictor
|
|
 |
TMHMM
Old hidden Markov model for membrane protein topology prediction, now superseded by Phobius as Phobius is better at all types of prediction.
|
|
 |
GPCRHMM
A hidden Markov model for GPCR detection
|
|
 |
SFINX
Multiple predictions of functional and structural features in proteins in an integrated graphical environment.
|
| |
Protein Domain Family Analysis |
|
 |
Pfam
A database of protein domain families.
Pfam is a large collection of multiple sequence alignments and hidden Markov models covering many common protein domains.
Try also
PfamAlyzer (under "Domain architecture").
|
|
 |
TreeDom
A flexible tool for analysing protein domain architecture evolution
|
|
 |
FunShift
A Database of Function Shift Analysis on Protein Subfamilies
|
|
 |
Belvu
Belvu is an X-windows viewer for multiple sequence alignments. It has an extensive set of modes to color the residues and features for fetching Swissprot entries and easy tracking of the position in the alignment.
|
|
 |
Stockholm
The "Stockholm" format is a system for marking up features in a multiple alignment. The Stockholm format is used by HMMER, Pfam, and Belvu.
|
 |
|
Other Analysis Tools |
|
 |
Dasher
A Java DAS client for displaying annotations on a protein sequence.
|
|
 |
MSP Crunch
A filtering tool for Blast matches.
|
|
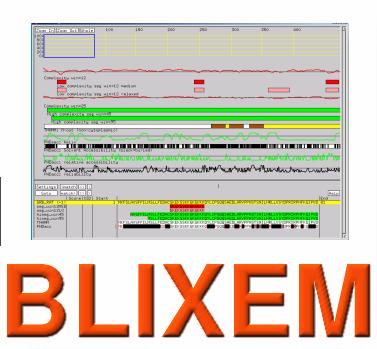 |
Blixem
Blixem, which stands for "BLast matches In an X-windows Embedded Multiple alignment", is an interactive browser of pairwise Blast matches that have been stacked up in a "master-slave" multiple alignment.
|
|
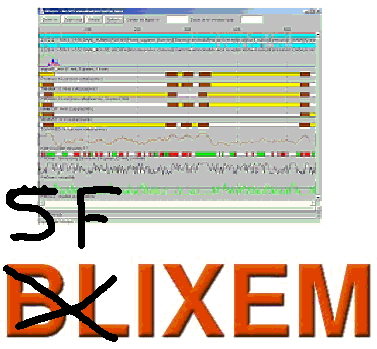 |
Sfixem
A java version of Blixem primarily designed for displaying SFS data. Handles HSPs, GFF, segment and curve data generated by various tools available on this server and elsewhere.
|
|
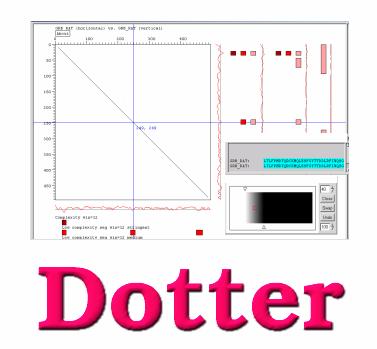 |
Dotter
A dot-matrix program with interactive greyscale rendering for genomic DNA and Protein sequence analysis.
|
|
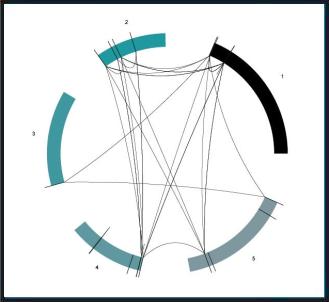 |
Chromowheel
ChromeWheel is a processing method for generating interactive illustrations of genome data. With this method chromosomes, genes and relations between these genes is displayed. The chromosomes are placed in a circle to avoid lines representing relations crossing genes and chromosomes.
|
 |
|
Multiple Sequence Alignments Tools |
|
 |
Belvu
Belvu - View multiple alignments in good-looking colours
|
|
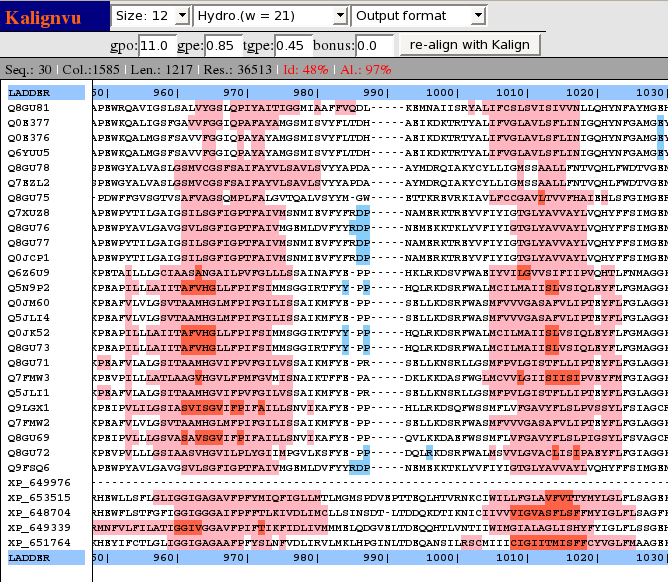 |
Kalign
Kalign - fast and accurate multiple alignments
|
 |
|
siRNA Design Tools |
 |
|
To list our repository of programs and datafiles,
Click Here.
|